PREDIPATH : analysis of the 65x65 data
Clickable table of contents
1. Description of the data
2. Analysis of the 0/1 data: reduction of dimension
3. Analysis of the 0/1 data: prediction of the status
4. Analysis of the percentage data: reduction of dimension
5. Analysis of the percentage data: prediction of the status
6. Concluding remarks
1. Description of the data
The original data is a 65x65 matrix of percentages of identity.
The genomes, described in the file allGroups are known to be commensal or pathogenic, as shown in the following excerpt:
ASSEMBLY_GENOME SPECIES GROUP
GCF_000026185.1 Erwinia_tasmaniensis_Et1 commensal
GCF_000196615.1 Erwinia_billingiae_Eb661|Eb661 commensal
GCF_000336255.1 Erwinia_toletana_DAPP-PG_735|DAPP-PG_735 commensal
GCF_000745075.1 Erwinia_sp._9145|9145 commensal
GCF_000770305.1 Erwinia_oleae|DAPP-PG531 commensal
GCF_000773975.1 Erwinia_typographi|M043b commensal
[...]
GCF_002732435.1 Erwinia_amylovora|NHWL02-2 pathogenic
GCF_002732445.1 Erwinia_amylovora|OR1 pathogenic
GCF_002732485.1 Erwinia_amylovora|MANB02-1 pathogenic
GCF_002732505.1 Erwinia_amylovora|EA110 pathogenic
GCF_002803865.1 Erwinia_amylovora|E-2 pathogenic
GCF_002952315.1 Erwinia_pyrifoliae|EpK1 pathogenic
The clusters are described in the file allClusters. Some clusters are shown below:
Name Assembly_genome
cluster_1 argannot~~(Bla)Penicillin_Binding_Protein_Ecoli:CP002291:664439-666340:1902
cluster_2 card~~gb|AAC75733.1|ARO:3000074|emrB
cluster_3 card~~gb|AIL15701|ARO:3003775|Escherichia
cluster_4 card~~gb|BAE77595.1|ARO:3003303|Escherichia
...
cluster_64 vfdb_pre~~VFG043692(gi:112292717)
cluster_65 vfdb_pre~~VFG044340(gi:292489703)
In order to better see the information, cluster names have been shortened and normalized: cluster_x becomes CL0x. The names of the genomes have been shortened too and prefixed by C for commensal or by P for pathogenic. Here is a part of the data to be analyzed, the complete file is here:
Short-name status CL01 CL02 CL03 CL04 CL05 CL06 CL07 CL08 CL09 CL10 CL11 CL12 CL13 CL14 CL15
C-0026185.1 commensal 0 0 0 0 98.095 0 0 0 0 0 95.976 0 90.984 0.00 90.783
C-0196615.1 commensal 0 0 0 0 99.048 0 0 0 0 0 95.380 0 90.164 0.00 0.000
C-0336255.1 commensal 0 0 0 0 98.571 0 0 0 0 0 95.746 0 90.164 90.84 90.668
C-0745075.1 commensal 0 0 0 0 98.571 0 0 0 0 0 96.051 0 0.000 0.00 91.129
C-0770305.1 commensal 0 0 0 0 98.571 0 0 0 0 0 96.051 0 0.000 0.00 91.129
[...]
P-2732445.1 pathogenic 0 0 0 0 98.571 0 0 0 0 0 96.051 0 0 0 0.000
P-2732485.1 pathogenic 0 0 0 0 98.095 0 0 0 0 0 96.051 0 0 0 0.000
P-2732505.1 pathogenic 0 0 0 0 98.571 0 0 0 0 0 95.976 0 0 0 0.000
P-2803865.1 pathogenic 0 0 0 0 98.571 0 0 0 0 0 96.051 0 0 0 0.000
P-2952315.1 pathogenic 0 0 0 0 98.571 0 0 0 0 0 95.753 0 0 0 90.323
2. Analysis of the 0/1 data: reduction of dimension
The original data have been converted to 0/1 data: if the percentage is not 0 then the value is 1. This provides a matrix of presence/absence to be analyzed.
Here is a part of the data to be analyzed, the complete file is here in text format and there in CSV format:
Genome status CL01 CL02 CL03 CL04 CL05 CL06 CL07 CL08 CL09 CL10 CL11 CL12 CL13 CL14 CL15
C-0026185.1 commensal 0 0 0 0 1 0 0 0 0 0 1 0 1 0 1
C-0196615.1 commensal 0 0 0 0 1 0 0 0 0 0 1 0 1 0 0
C-0336255.1 commensal 0 0 0 0 1 0 0 0 0 0 1 0 1 1 1
C-0745075.1 commensal 0 0 0 0 1 0 0 0 0 0 1 0 0 0 1
C-0770305.1 commensal 0 0 0 0 1 0 0 0 0 0 1 0 0 0 1
[...]
P-2732445.1 pathogenic 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0
P-2732485.1 pathogenic 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0
P-2732505.1 pathogenic 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0
P-2803865.1 pathogenic 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0
P-2952315.1 pathogenic 0 0 0 0 1 0 0 0 0 0 1 0 0 0 1
As one can see, some columns look similar. And in fact, some colums are equal. So we first wrote a script to remove them. Here is the result of this script:
NB Column Equal-to:
01 status
02 CL01 CL02 CL04 CL06 CL07 CL08 CL09 CL10 CL12 CL23 CL53 CL55
04 CL03
06 CL05
12 CL11
14 CL13
15 CL14
16 CL15
17 CL16
18 CL17 CL52
19 CL18
20 CL19 CL20
22 CL21
23 CL22
25 CL24
26 CL25
27 CL26 CL40
28 CL27
29 CL28 CL29 CL31 CL32 CL38
31 CL30
34 CL33 CL39 CL42 CL44 CL46 CL47
35 CL34 CL36 CL49 CL65
36 CL35 CL50
38 CL37
42 CL41
44 CL43
46 CL45
49 CL48
52 CL51
55 CL54
57 CL56 CL57 CL58 CL59 CL60 CL61 CL62 CL63 CL64
So instead of analyzing 65 columns of 0/1 data, we have to analyze only 30 columns. Since no resulting column if constant (details not shown here), we can proceed to predict the status of the genomes.
3. Analysis of the 0/1 data: prediction of the status
The classical statistical method for the classification of two classes is called binary logistic regression. Here it fails since there is a complete separation of the data, which means there is a combination of some columns that predicts exactly the status.
We have found such a combination (probably minimal) with 4 columns:
Coefficients Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.03273 0.02297 44.961 < 2e-16 ***
CL41 0.96727 0.05544 17.449 < 2e-16 ***
CL33 0.45818 0.06591 6.952 3.02e-09 ***
CL34 -0.45818 0.08175 -5.605 5.56e-07 ***
CL13 -0.08364 0.03574 -2.340 0.0226 *
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.08739 on 60 degrees of freedom
Multiple R-squared: 0.9697, Adjusted R-squared: 0.9677
F-statistic: 480.6 on 4 and 60 DF, p-value: < 2.2e-16
Comparing status and round(ml$fitted.values),0): perfect separation!
The minimal data needed to predict the status is then shown below:
Details of the predictors values
================================
Status 1 = commensal
status CL41 CL33 CL34 CL13
C-0026185.1 1 1 0 1 1
C-0196615.1 1 0 0 0 1
C-0336255.1 1 0 0 0 1
C-0745075.1 1 0 0 0 0
C-0770305.1 1 0 0 0 0
C-0773975.1 1 0 0 0 0
C-1267535.1 1 0 0 0 1
C-1267545.1 1 0 0 0 1
C-1269445.1 1 0 0 0 1
C-1422605.1 1 0 0 0 0
C-1484765.1 1 0 0 0 1
C-1517405.1 1 0 0 0 0
C-1571305.1 1 0 0 0 1
C-2751995.1 1 0 0 0 0
C-2752015.1 1 0 0 0 0
C-2752025.1 1 0 0 0 0
C-2752055.1 1 0 0 0 0
C-2752075.1 1 0 0 0 0
C-2752165.1 1 0 0 0 0
C-2752575.1 1 0 0 0 1
C-2752595.1 1 0 0 0 0
C-2865965.1 1 0 0 0 0
C-2980095.1 1 0 0 0 1
C-0068895.1 1 0 0 0 0
Status 2 = pathogenic
status CL41 CL33 CL34 CL13
P-0026985.1 2 1 1 1 0
P-0027205.1 2 1 1 1 0
P-0027265.1 2 1 1 1 0
P-0091565.1 2 1 1 1 0
P-0165815.1 2 1 1 1 0
P-0240705.2 2 1 1 1 0
P-0367545.1 2 1 1 1 0
P-0367565.1 2 1 1 1 0
P-0367585.1 2 1 1 1 0
P-0367605.1 2 1 1 1 0
P-0367625.1 2 1 1 1 0
P-0367645.1 2 1 1 1 0
P-0367665.1 2 1 1 1 0
P-0404125.1 2 1 0 0 0
P-0513355.1 2 1 1 1 0
P-0513395.1 2 1 1 1 0
P-0513415.1 2 1 1 1 0
P-0590885.1 2 1 0 0 0
P-0696075.1 2 1 1 1 0
P-0975275.1 2 1 0 0 0
P-1050515.1 2 1 0 1 0
P-2732125.1 2 1 1 1 0
P-2732175.1 2 1 1 1 0
P-2732205.1 2 1 1 1 0
P-2732215.1 2 1 1 1 0
P-2732245.1 2 1 1 1 0
P-2732255.1 2 1 1 1 0
P-2732285.1 2 1 1 1 0
P-2732295.1 2 1 1 1 0
P-2732315.1 2 1 1 1 0
P-2732335.1 2 1 1 1 0
P-2732365.1 2 1 1 1 0
P-2732385.1 2 1 1 1 0
P-2732405.1 2 1 1 1 0
P-2732425.1 2 1 1 1 0
P-2732435.1 2 1 1 1 0
P-2732445.1 2 1 1 1 0
P-2732485.1 2 1 1 1 0
P-2732505.1 2 1 1 1 0
P-2803865.1 2 1 1 1 0
P-2952315.1 2 1 1 1 0
Since these data correspond to presence/absence, a count-heatmap is probably a better way to understand these data (you can click on the image to have a better view):

Columns: CL13 CL41 CL33 CL34; Rows: commensal then pathogenic.
Since the linear regression model is not easy to understand, the main columns used to predict the status are given below but none is specific, which means that alone none is sufficient for the prediction:
Thresholds : presence : pct1 >= 90 %; absence: pct0 < 10 %
no cluster is able to predict "commensal"
6 clusters nearly allow to predict "pathogenic"
NB Column sum sumCommens pctCommens sumPatho pctPatho specCommens specPatho presqueCommens presquePatho
19 CL33 37 0 0.00 37 90.24 YYY
20 CL34 39 1 4.17 38 92.68 YYY
21 CL35 40 1 4.17 39 95.12 YYY
23 CL41 42 1 4.17 41 100.00 YES YYY
24 CL43 38 0 0.00 38 92.68 YYY
25 CL45 41 1 4.17 40 97.56 YYY
Details of the predicted values:
================================
NB Column sum sumCommens pctCommens sumPatho pctPatho specCommens specPatho presqueCommens presquePatho
01 CL01 1 1 4.17 0 0.00
02 CL03 4 4 16.67 0 0.00
03 CL11 64 23 95.83 41 100.00 YES XXX YYY
04 CL13 10 10 41.67 0 0.00
05 CL14 12 12 50.00 0 0.00
06 CL15 29 21 87.50 8 19.51
07 CL16 6 6 25.00 0 0.00
08 CL17 2 2 8.33 0 0.00
09 CL18 44 6 25.00 38 92.68 YYY
10 CL19 1 0 0.00 1 2.44
11 CL21 62 23 95.83 39 95.12 XXX YYY
12 CL22 1 1 4.17 0 0.00
13 CL24 14 0 0.00 14 34.15
14 CL25 30 0 0.00 30 73.17
15 CL26 32 0 0.00 32 78.05
16 CL27 30 0 0.00 30 73.17
17 CL28 33 0 0.00 33 80.49
18 CL30 29 0 0.00 29 70.73
19 CL33 37 0 0.00 37 90.24 YYY
20 CL34 39 1 4.17 38 92.68 YYY
21 CL35 40 1 4.17 39 95.12 YYY
22 CL37 34 0 0.00 34 82.93
23 CL41 42 1 4.17 41 100.00 YES YYY
24 CL43 38 0 0.00 38 92.68 YYY
25 CL45 41 1 4.17 40 97.56 YYY
26 CL48 36 0 0.00 36 87.80
27 CL51 2 0 0.00 2 4.88
28 CL54 2 2 8.33 0 0.00
29 CL56 1 1 4.17 0 0.00
So, roughly speaking, cluster 41 predicts nearly pathogenicity. The three other clusters are here to take care of genome C-0026185.1.
Genome status predit arrondi
C-0026185.1 1 1.4581818 1
C-0196615.1 1 0.9490909 1
C-0336255.1 1 0.9490909 1
C-0745075.1 1 1.0327273 1
C-0770305.1 1 1.0327273 1
C-0773975.1 1 1.0327273 1
C-1267535.1 1 0.9490909 1
C-1267545.1 1 0.9490909 1
C-1269445.1 1 0.9490909 1
C-1422605.1 1 1.0327273 1
C-1484765.1 1 0.9490909 1
C-1517405.1 1 1.0327273 1
C-1571305.1 1 0.9490909 1
C-2751995.1 1 1.0327273 1
C-2752015.1 1 1.0327273 1
C-2752025.1 1 1.0327273 1
C-2752055.1 1 1.0327273 1
C-2752075.1 1 1.0327273 1
C-2752165.1 1 1.0327273 1
C-2752575.1 1 0.9490909 1
C-2752595.1 1 1.0327273 1
C-2865965.1 1 1.0327273 1
C-2980095.1 1 0.9490909 1
C-0068895.1 1 1.0327273 1
P-0026985.1 2 2.0000000 2
P-0027205.1 2 2.0000000 2
P-0027265.1 2 2.0000000 2
P-0091565.1 2 2.0000000 2
P-0165815.1 2 2.0000000 2
P-0240705.2 2 2.0000000 2
P-0367545.1 2 2.0000000 2
P-0367565.1 2 2.0000000 2
P-0367585.1 2 2.0000000 2
P-0367605.1 2 2.0000000 2
P-0367625.1 2 2.0000000 2
P-0367645.1 2 2.0000000 2
P-0367665.1 2 2.0000000 2
P-0404125.1 2 2.0000000 2
P-0513355.1 2 2.0000000 2
P-0513395.1 2 2.0000000 2
P-0513415.1 2 2.0000000 2
P-0590885.1 2 2.0000000 2
P-0696075.1 2 2.0000000 2
P-0975275.1 2 2.0000000 2
P-1050515.1 2 1.5418182 2
P-2732125.1 2 2.0000000 2
P-2732175.1 2 2.0000000 2
P-2732205.1 2 2.0000000 2
P-2732215.1 2 2.0000000 2
P-2732245.1 2 2.0000000 2
P-2732255.1 2 2.0000000 2
P-2732285.1 2 2.0000000 2
P-2732295.1 2 2.0000000 2
P-2732315.1 2 2.0000000 2
P-2732335.1 2 2.0000000 2
P-2732365.1 2 2.0000000 2
P-2732385.1 2 2.0000000 2
P-2732405.1 2 2.0000000 2
P-2732425.1 2 2.0000000 2
P-2732435.1 2 2.0000000 2
P-2732445.1 2 2.0000000 2
P-2732485.1 2 2.0000000 2
P-2732505.1 2 2.0000000 2
P-2803865.1 2 2.0000000 2
P-2952315.1 2 2.0000000 2
The predictions are not robust since there are some values around 1.5; it is difficult to assign them to class 1 or 2.
4. Analysis of the percentage data: reduction of dimension
So now we are using the original data (percentages of identity). As before, we try to remove equal columns. This time, only one column can be removed:
NB Column Equal-to:
01 status
02 CL01
03 CL02
04 CL03
05 CL04
06 CL05
07 CL06
08 CL07
09 CL08
10 CL09
11 CL10
12 CL11
13 CL12
14 CL13
15 CL14
16 CL15
17 CL16
18 CL17
19 CL18
20 CL19 CL20
22 CL21
23 CL22
24 CL23
25 CL24
26 CL25
27 CL26
28 CL27
29 CL28
30 CL29
31 CL30
32 CL31
33 CL32
34 CL33
35 CL34
36 CL35
37 CL36
38 CL37
39 CL38
40 CL39
41 CL40
42 CL41
43 CL42
44 CL43
45 CL44
46 CL45
47 CL46
48 CL47
49 CL48
50 CL49
51 CL50
52 CL51
53 CL52
54 CL53
55 CL54
56 CL55
57 CL56
58 CL57
59 CL58
60 CL59
61 CL60
62 CL61
63 CL62
64 CL63
65 CL64
5. Analysis of the percentage data: prediction of the status
Here again the binary logistic regression fails since there is also a complete separation of the data, which means there is a combination of some columns that performs a complete separation of the data with respect to the status.
We have probably found a minimal solution with these 3 columns and their combination:
Coefficients Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.0001358 0.0187541 53.329 < 2e-16 ***
CL41 0.0108103 0.0005939 18.201 < 2e-16 ***
CL33 0.0046968 0.0006854 6.853 4.14e-09 ***
CL34 -0.0054701 0.0008640 -6.331 3.22e-08 ***
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.08994 on 61 degrees of freedom
Multiple R-squared: 0.9674, Adjusted R-squared: 0.9658
F-statistic: 603.5 on 3 and 61 DF, p-value: < 2.2e-16
Comparing status and round(ml$fitted.values),0): perfect separation!
The minimal data needed to predict the status is shown below:
Equal values
============
genome status CL33 CL34 CL41
C-0196615.1 1 0 0 0
C-0336255.1 1 0 0 0
C-0745075.1 1 0 0 0
...
P-2732485.1 2 100 100 100
P-2732505.1 2 100 100 100
P-2803865.1 2 100 100 100
Unequal values
==============
genome status CL33 CL34 CL41
C-0026185.1 1 0 97 94
P-0026985.1 2 92 98 99
P-0027265.1 2 92 98 99
P-0165815.1 2 92 98 99
P-0404125.1 2 0 0 92
P-0590885.1 2 0 0 94
P-0975275.1 2 0 0 92
P-1050515.1 2 0 94 94
P-2952315.1 2 92 98 99
Here are the predicted values:
genome status predicted rounded
C-0026185.1 1 1.490340 1
C-0196615.1 1 1.000136 1
C-0336255.1 1 1.000136 1
C-0745075.1 1 1.000136 1
C-0770305.1 1 1.000136 1
C-0773975.1 1 1.000136 1
C-1267535.1 1 1.000136 1
C-1267545.1 1 1.000136 1
C-1269445.1 1 1.000136 1
C-1422605.1 1 1.000136 1
C-1484765.1 1 1.000136 1
C-1517405.1 1 1.000136 1
C-1571305.1 1 1.000136 1
C-2751995.1 1 1.000136 1
C-2752015.1 1 1.000136 1
C-2752025.1 1 1.000136 1
C-2752055.1 1 1.000136 1
C-2752075.1 1 1.000136 1
C-2752165.1 1 1.000136 1
C-2752575.1 1 1.000136 1
C-2752595.1 1 1.000136 1
C-2865965.1 1 1.000136 1
C-2980095.1 1 1.000136 1
C-0068895.1 1 1.000136 1
P-0026985.1 2 1.965778 2
P-0027205.1 2 2.003826 2
P-0027265.1 2 1.965778 2
P-0091565.1 2 2.003826 2
P-0165815.1 2 1.965778 2
P-0240705.2 2 2.003826 2
P-0367545.1 2 2.003826 2
P-0367565.1 2 2.003826 2
P-0367585.1 2 2.003826 2
P-0367605.1 2 2.003826 2
P-0367625.1 2 2.003826 2
P-0367645.1 2 2.003826 2
P-0367665.1 2 2.003826 2
P-0404125.1 2 1.999958 2
P-0513355.1 2 2.003826 2
P-0513395.1 2 2.003826 2
P-0513415.1 2 2.003826 2
P-0590885.1 2 2.014876 2
P-0696075.1 2 2.003826 2
P-0975275.1 2 1.999958 2
P-1050515.1 2 1.502364 2
P-2732125.1 2 2.003826 2
P-2732175.1 2 2.003826 2
P-2732205.1 2 2.003826 2
P-2732215.1 2 2.003826 2
P-2732245.1 2 2.003826 2
P-2732255.1 2 2.003826 2
P-2732285.1 2 2.003826 2
P-2732295.1 2 2.003826 2
P-2732315.1 2 2.003826 2
P-2732335.1 2 2.003826 2
P-2732365.1 2 2.003826 2
P-2732385.1 2 2.003826 2
P-2732405.1 2 2.003826 2
P-2732425.1 2 2.003826 2
P-2732435.1 2 2.003826 2
P-2732445.1 2 2.003826 2
P-2732485.1 2 2.003826 2
P-2732505.1 2 2.003826 2
P-2803865.1 2 2.003826 2
P-2952315.1 2 1.965778 2
Here also a count-heatmap is probably a better way to understand these data (you can click on the image to have a better view):
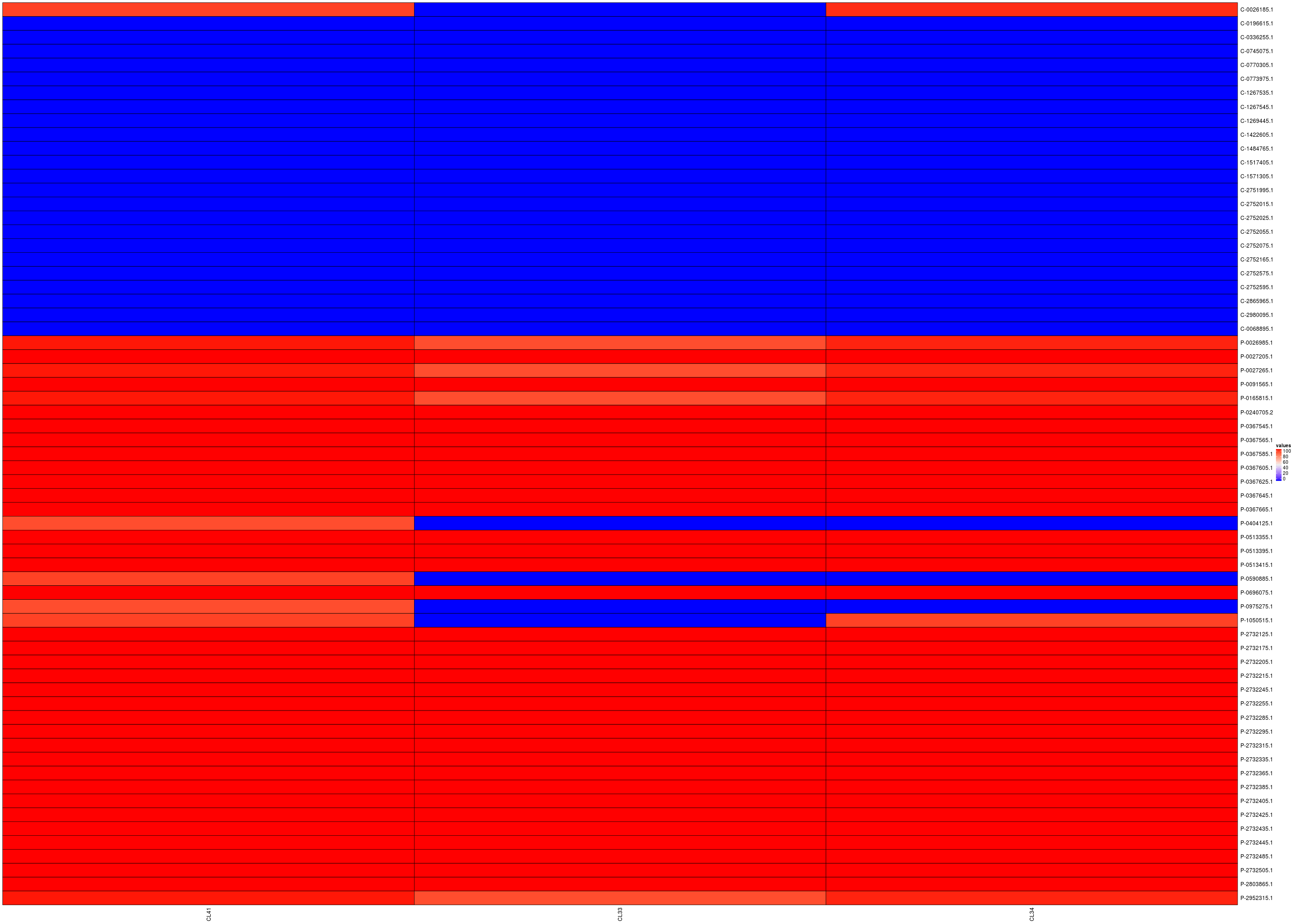
Columns: CL41 CL33 CL34; Rows: commensal then pathogenic.
So, as previously, cluster 41, named vfdb_pre~~VFG041501(gi:292487026), is nearly a predictor of pathogenicity since all pathogenic genomes have a non zero value for this cluster and all commensal genomes are null for this cluster except the genome named GCF_000026185.1 Erwinia_tasmaniensis_Et.
As before the predictions are not robust since again there are some values around 1.5; it is also difficult to assign them to class 1 or 2.
6. Concluding remarks
Though we have found some predictors for the status, the solution is not robust since one genomes can change it all.
If it was possible to ignore genome C-0026185.1 the predictor for pathogenicity would be exactly the presence of cluster 41, identified as vfdb_pre~~VFG041501(gi:292487026).
One has to think, using the 0/1 matrix, of the clusters equal to the predictors CL33 and CL34.
In order to get a robust classification, we have to think of a new strategy:
-
We can remove genome named GCF_000026185.1 Erwinia_tasmaniensis_Et so pathogenicity can be considered as equal to CL41 greater than 90.
-
We can analyze the real status of genome named GCF_000026185.1 Erwinia_tasmaniensis_Et for it may be pathogenic with a bad annotation.
-
We can decide to add more data to be sure of the information.
-
We can refine pathogenicity using more than two levels.
Code-source de cette page.
|  Retour à la page principale de
(gH)
Retour à la page principale de
(gH)

